This assignment is to be done either individually or in pairs. Do not show your code to any other group and do not look at any other group's code. Do not put your code in a public directory or otherwise make it public. However, you may get help from the TAs or the instructor. You are encouraged to use the LMS Discussions page to post problems so that other students can also answer/see the answers.
In this programming assignment, you will implement a concurrent and distributed DNA Sequence Matching counter. The counter should take two DNA sequences: an input sequence which will be grouped together by the chemical bases that would be present in a target sequence. The program should return the total number of inversions we need to perform for a sequence match between the input and target sequences. Your assignment should be created using the actor model to allow for concurrent and distributed computation of the number of inversions needed to match the target DNA sequence.
Note : The DNA sequence is made up of four chemical bases: adenine (a), guanine (g), cytosine (c), and thymine (t).
An inversion is a pair of consecutive places of a sequence where the elements on these places are out of their natural order. Inversion count is defined by the total number of inversions needed to convert an input to a target sequence.
| Target Seq | Input Seq | Inversion Count | Inversions |
|---|---|---|---|
| ga | ga | 0 | - |
| ga | ag | 1 | [(a,g)] |
| gat | tga | 2 | [(t,g), (t,a)] |
| gat | tag | 3 | [(t,a), (t,g), (g,a)] |
| gtca | tacg | 4 | [(g,c), (g,a), (g,t), (c,a)] |
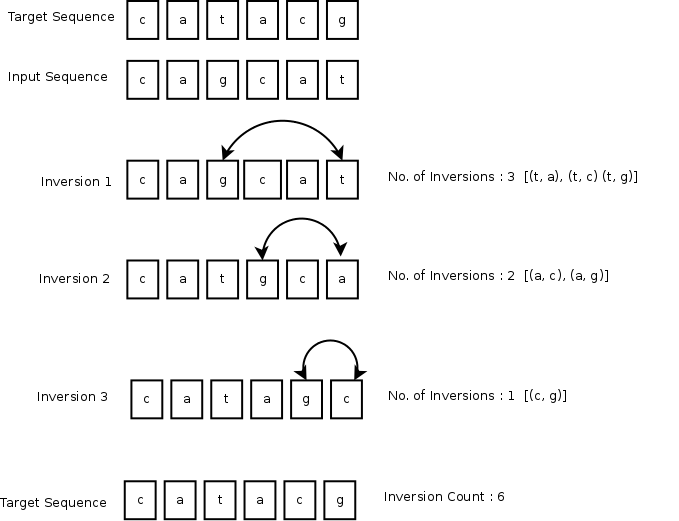
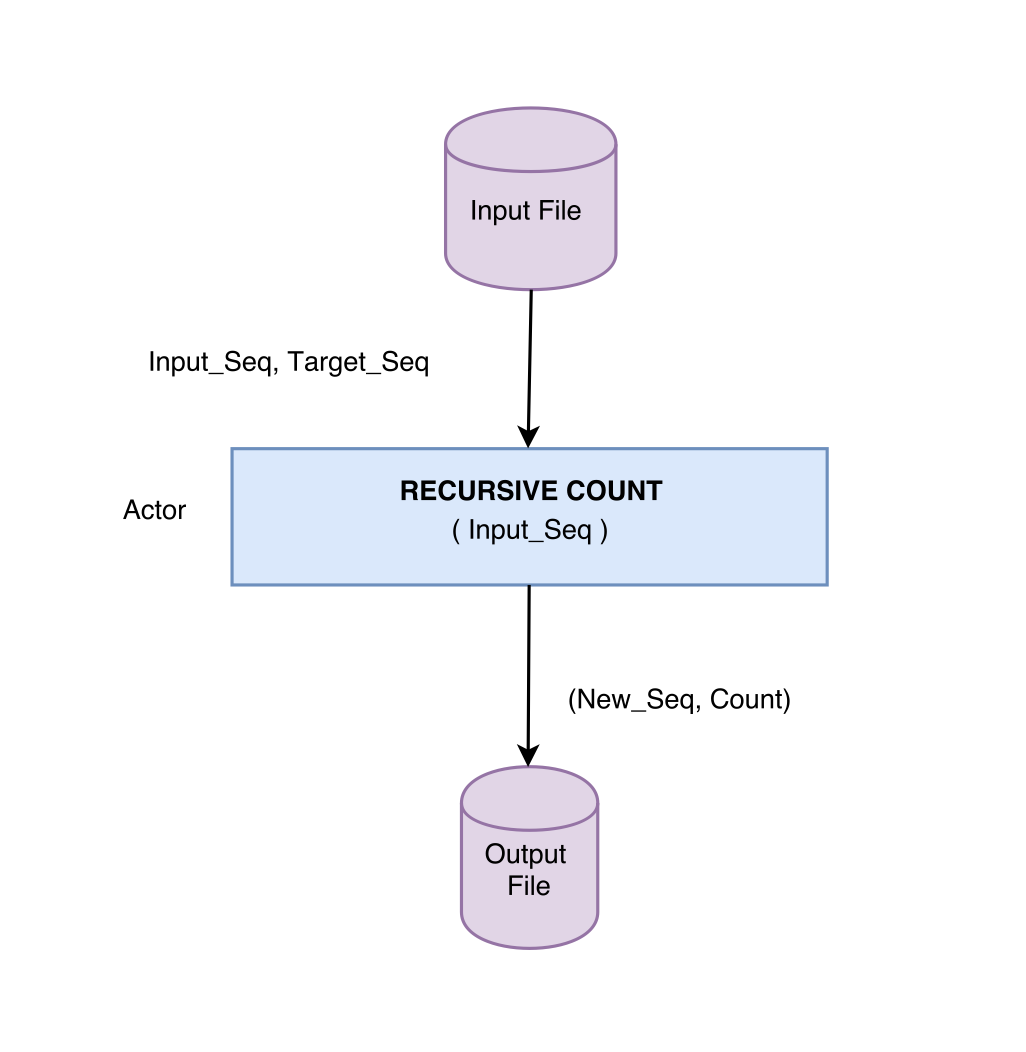
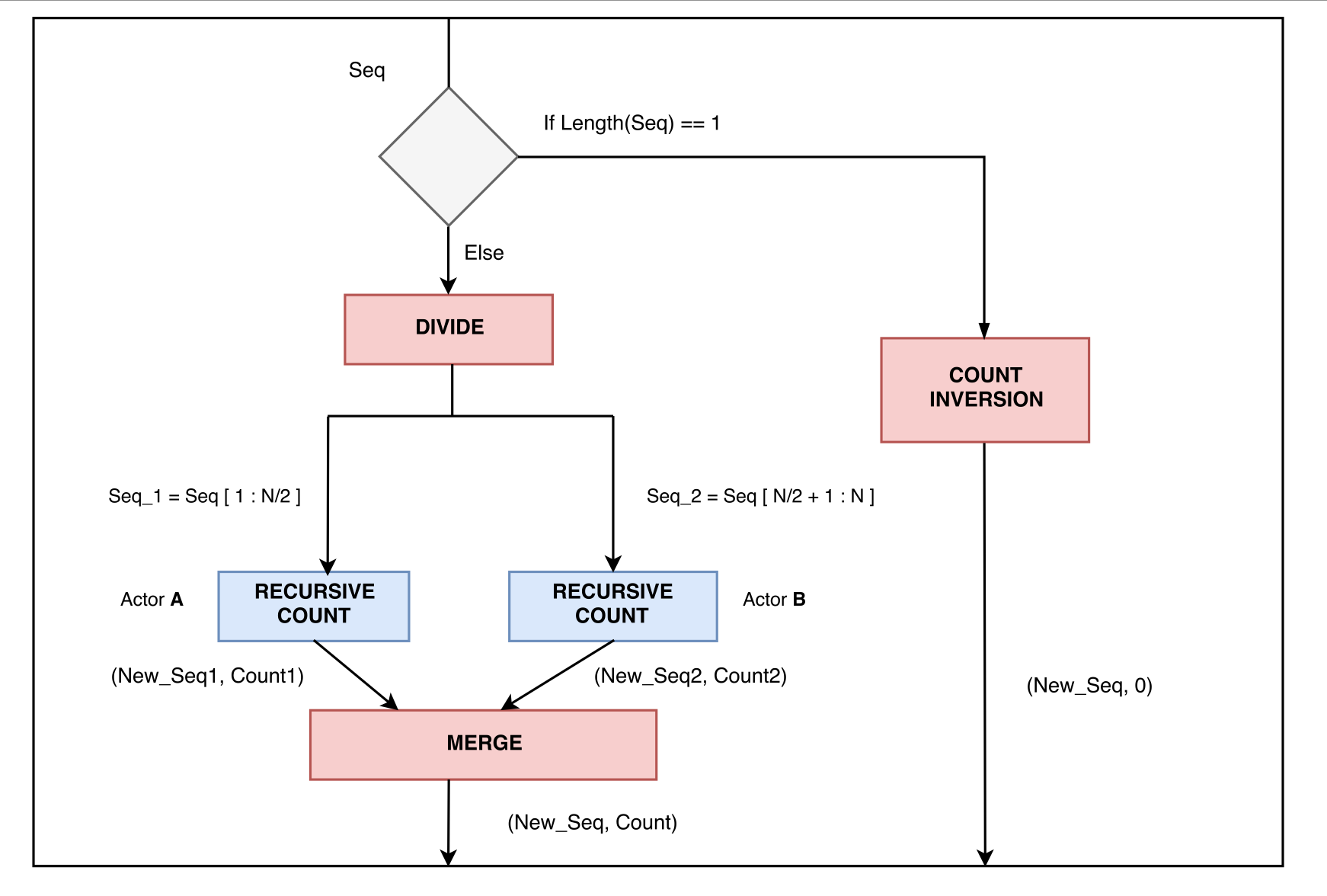
Hint: You can view inversion count as a modified sorting problem and use a correspondingly modified recursive merge-sort algorithm.
$ ./run.sh < ./input1.txt 0 $ ./run.sh < ./input2.txt 2 $ cat ./input1.txt ga ga $ cat ./input2.txt gaag agga
Your concurrent program should be run in the following manner:
$ salsac dna/* $ salsa dna.InversionCount input2.txt 2where
input2.txt specifies the name of input file, it should be read with the format specified above.
salsac and salsa are UNIX aliases or Windows
batch scripts that run java and javac with
the expected arguments: See .cshrc
for UNIX, and salsac.bat
salsa.bat
for Windows.
And your distributed program should be run in the following manner:
$ salsac dna/* $ salsa dna.DInversionCount input.txt theaters.txtwhere
input.txt specifies an input file name,
and theaters.txt is a name server and theater
description file.
See a sample name server and theaters description file, the first line of which specifies the name server location and each of the remaining lines specifies a theater location.
InversionCount behavior should be in a relative path dna/InversionCount.salsa, and should
start with the line module dna;.m1(...);m2(...); does not imply m1 occurs before m2.m(...)@n(...);, n is processed after m is executed, but not
necessarily after messages sent inside m are executed. For example, if inside m, messages
m1 and m2 are sent, in general, n could happen before m1 and m2.[host0:dir0]$ wwcns [port number 0] [host1:dir1]$ wwctheater [port number 1] [host2:dir2]$ wwctheater [port number 2] ...where
wwcns and wwctheater are UNIX aliases
or Windows batch scripts: See .cshrc
for UNIX, and wwcns.bat
wwctheater.bat
for Windows.
dna directory should be visible in directories:
host1:dir1 and host2:dir2.
Then, run the distributed program as mentioned above.
Please put the modified program in the file DInversionCount.salsa. Be sure to attempt to evenly distribute your workload across the nodes; a good way to do this is a random, uniform sampling of an array containing all nodes.
Please find the starting code on Github: Fall16_PA2_Erlang.
When submitting, make sure your main:start() launches your program. It should read two strings from the command line, and output the number of inversions it takes to make the first string into the second string.
machine1$ erl -sname example1@localhost -setcookie thecookiemonster machine2$ erl -sname example2@localhost -setcookie thecookiemonster machine3$ erl -sname example3@localhost -setcookie thecookiemonster machine4$ erl -sname example4@localhost -setcookie thecookiemonster -pa ./main.erl -run main -run init stop -noshell
You can use the following code to dynamically find available nodes:
get_random_node() -> rget_random_node(rand:uniform(length(net_adm:world())), net_adm:world()). rget_random_node(1, [H|_]) -> H; rget_random_node(_, [H|[]]) -> H; rget_random_node(N, [_|T]) -> rget_random_node(N-1, T).
Please look at http://erlang.org/doc/ for Erlang examples, function documentation, and usage. Pay special attention to http://erlang.org/doc/man/io.html, and remember Google is your friend (but don't copy and paste code!).
Due Date: Thursday, 10/27, 7:00PM
Grading: The assignment will be graded mostly on correctness, but code clarity / readability will also be a factor (comment, comment, comment!).
Submission Requirements: Please submit a ZIP file with your code, including a README file. README files must be in plain text; markdown is acceptable. Your ZIP file should be named with your LMS user name(s) and chosen language as the filename, either userid1_erlang.zip (or userid1_salsa.zip) or userid1_userid2_erlang.zip (or userid1_userid2_salsa.zip). Only submit one assignment per pair via LMS. In the README file, place the names of each group member (up to two). Your README file should also have a list of specific features / bugs in your solution.
Do not include unnecesary files. Test your archive after uploading it. Name your source files appropriately: InversionCount.salsa for SALSA, and main.erl for Erlang.